Grant Abstract: With our increasing ability to measure biological data at scale and the digitalization of health records, computational thinking is becoming ever more important in the biological science and healthcare. The research directions proposed in this grant look to build robust machine learning models and tools for computational biology by including principles and analysis from other engineering fields, like control, that have a proven record of incorporating robustness into the systems they have automated. This increased robustness will save resources during the development of these machine learning models. It will also lead to more reliable diagnostics, clinical tools, and machine learning based biological discoveries. We have proposed three future research directions at the intersection of machine learning, control, and computational biology (a) modeling dynamical systems, (b) robust optimization schemes (c) control principles for in vivo modeling of microbial communities. The first proposed research area involves the development of flexible models for performing inference on dynamical systems models with time-series data. Dynamical systems models are able to learn mathematically causal relationships between variables, compared to other models whose parameters may only have correlative relationships. Our flexible models will be differentiable allowing them to be trained using the same efficient algorithms and hardware that have propelled deep learning models into the spotlight. These differentiable methods will allow for us to more easily integrate the uncertainty associated with biological measurements into our models. The second research area looks to develop more robust gradient optimization algorithms, the work horse for training deep neural networks. Many of the popular algorithms used to train deep neural networks were not explicitly designed to be robust. By developing more robust optimization techniques machine learning models trained on disparate data sets at different hospitals or labs will be more reproducible and will require less time for tuning parameters, ultimately saving resources as well. These robust optimization techniques will also aid in the certification of machine learning based tools that will ultimately be deployed in the clinic. The third research area we propose is an approach for the discovery and design of robust microbial communities. Communities of commensal, or engineered, bacteria have long been proposed as alternative therapies for the treatment of gut related illness (“bugs as drugs”). We propose a top down approach to identifying putative microbial consortia members from time-series experiments with germ free mice colonized by complex flora. By beginning the consortia design process in vivo we hope to overcome the challenge that many other attempts at consortia construction have encountered where in vitro designed communities do not reproduce their intended properties once transferred into living host organisms. The tools from this work will be built using open access software and all data will be made easily accessible and explorable to the public.

Gibson Lab receives $450K NIH R21 grant “Tracking the microbiome: purpose-built machine learning tools for tracking microbial strains over time”
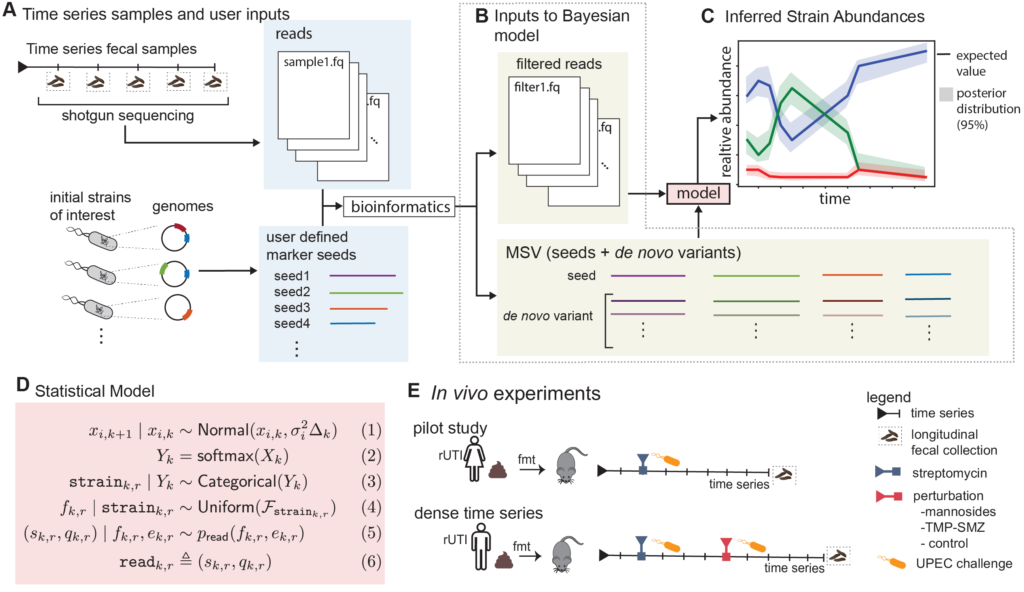
Grant Abstract: Approximately 150 million people annually experience urinary tract infections (UTI), the most common cause of which is uropathogenic Escherichia coli (UPEC). The gut is a known reservoir of UPEC, which typically reside at low abundance, but can transcend the periurethral area to invade the bladder. While the E. coli population within the gut can be diverse, it has been suggested that certain strains have a greater propensity to migrate and cause infection. This may be one driving factor to explain why half of those with an acute infection have a recurrence even after taking antibiotics that clear the first infection from the urinary tract. Being able to detect and track E. coli strains over time would have direct clinical applications for those patients who have frequent recurrences due to gut UPEC carriage. One such clinical application would be early detection and intervention before the onset of infection. Unfortunately, current metagenomic algorithms are not capable of performing strain tracking accurately enough for clinical relevance, especially for low abundance species such as E. coli. A major factor for this lack of accuracy is that all current state-of-the-art metagenomic tools completely ignore temporal dependence between samples. Even if it is known that multiple samples are from the same patient, current tools analyze those samples as if they were independent. Furthermore, many metagenomic tools ignore the sequence quality information that is provided for every nucleobase in every read. We propose to develop a more precise strain tracking algorithm that does take this additional information into account, making the tool host-time-quality aware. Finally, we will pilot and validate our algorithm on a clinically relevant gnotobiotic colonization model. Specifically, humanized germ-free mice will be undergoing two rounds of E. coli challenges with therapeutic perturbations from antibiotics or mannosides, a small molecule precision antibiotic-sparing therapeutic. We propose the following specific aims: (1) Develop the first purpose-built computational method for tracking bacterial strains in the microbiome over time, (2) Gnotobiotic mouse model undergoing UPEC challenges and a therapeutic perturbation. These aims would advance the microbiome field forward allowing for the future development of therapeutics and clinical diagnostics.
Link to NIH Award – Gibson Lab Website