BWH COMPUTATIONAL PATHOLOGY
Our Mission
Improve human health
Our core mission is to alleviate human suffering by reducing the burden of diseases on individuals and on the population. This mission informs all our activities in developing and applying computational technologies, which we leverage to make an impact on a broad range of human diseases including infectious, cancer, heart, kidney, intestinal, autoimmune, allergies, and neurological disorders.
Advance the field of pathology
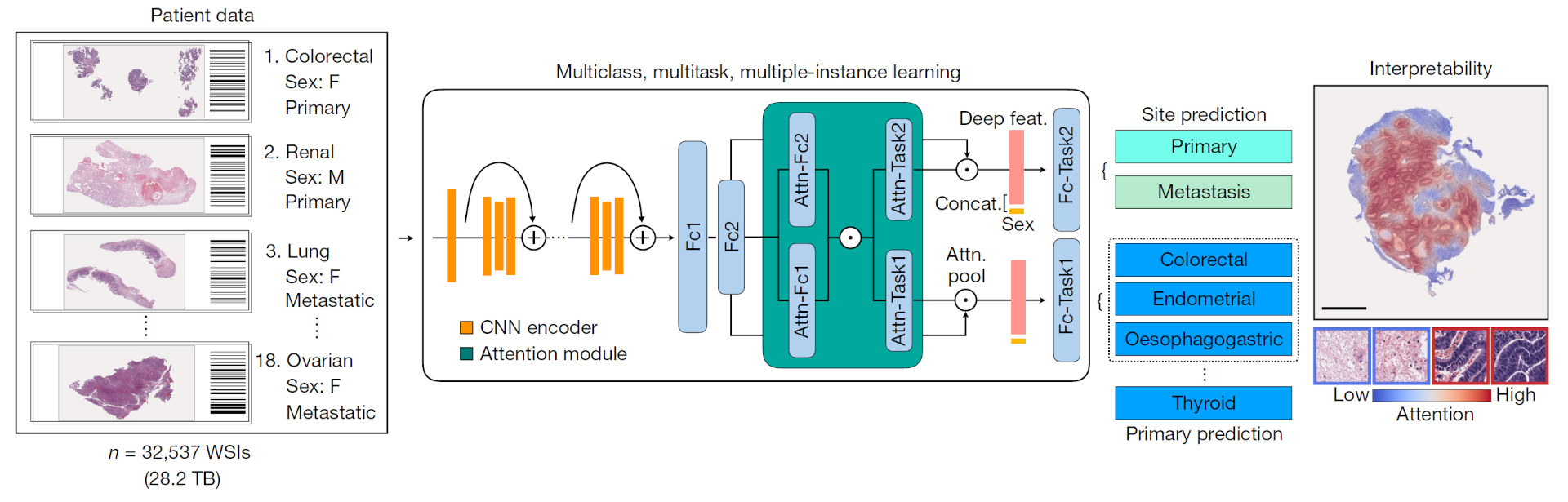
Pathology is both a scientific and a medical discipline, involving the study of basic mechanisms of diseases and the diagnosis of diseases using tissue and fluid samples. Our goal is to advance the field of pathology through computational technologies, to improve the understanding, diagnosis, and treatment of human diseases. With this broad view of pathology, we work on a variety of applications such as deep learning/artificial intelligence to improve cancer diagnosis/prognosis from histology images and to create new live bacterial therapeutics to treat infectious or autoimmune diseases.
Develop innovative computational methods
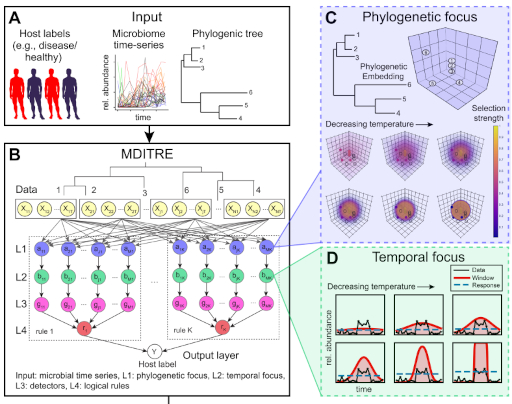
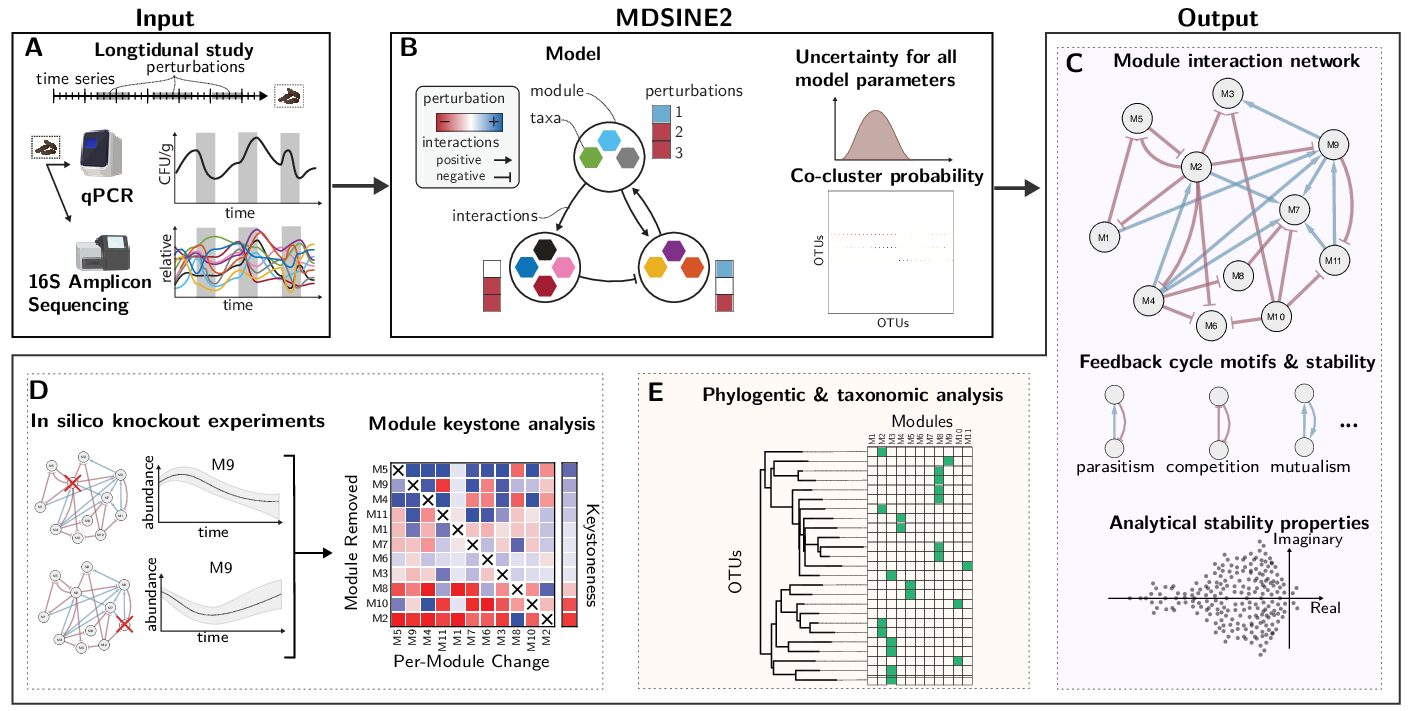
Human diseases often have complex causes and effects on the body. Data needed to analyze human diseases is similarly complex and multi-faceted. These data are also often difficult to acquire, leading to relatively limited dataset sizes. These and other challenges necessitate going beyond application of existing computational methods. Thus, we actively engage in computational research, to develop novel computational models, inference algorithms, integrated pipelines, and hardware. To accomplish this, we leverage a variety of advanced computational disciplines, including Bayesian nonparametric statistics, deep learning and control theory.
Division Opportunities
Division News
-
Dan MacDonald awarded Banting Fellowship
Daniel MacDonald, a Research Fellow in the Gibson Lab, is a 2024 recipient of the Banting Postdoctoral Fellowship. The Banting Postdoctoral Fellowship program provides funding to the very best postdoctoral […]
-
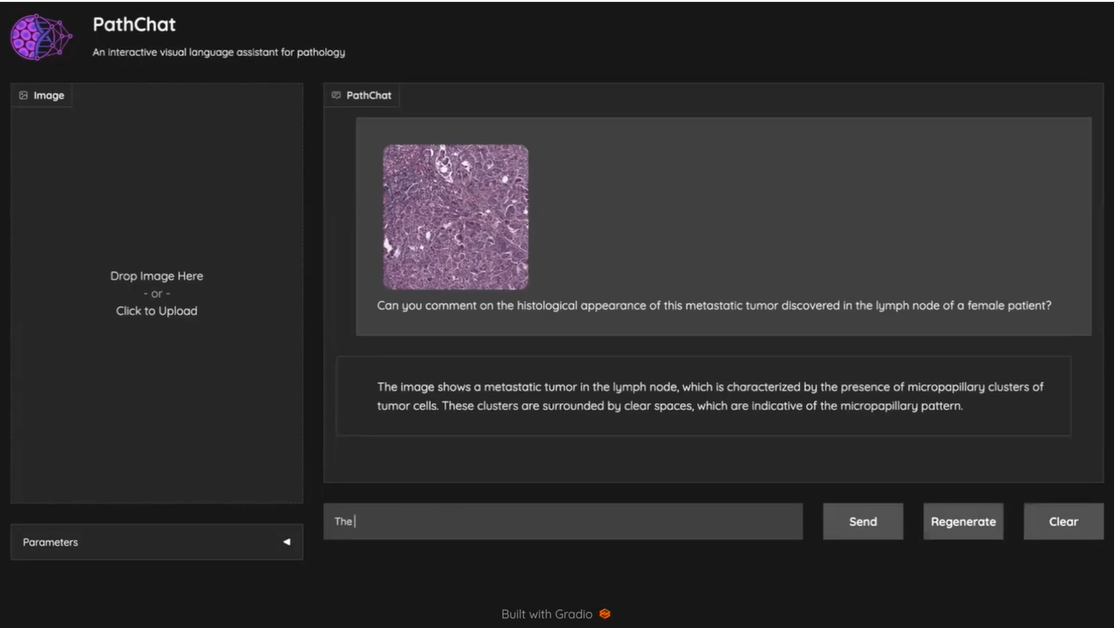
Mahmood Lab releases PathChat, a vision-language AI assistant for Pathology
Developed by the Mahmood Lab, PathChat is a vision-language AI assistant for Pathology that can analyze histology images and answer diverse pathology-related queries. See a demonstration of PathChat. The PathChat publication […]
-
May ABC Seminar: Jean du Terrail, PhD – Owkin – “Federated Learning in Healthcare in the Real-World: Examples and Practical Challenges”
With the never-ending revolutions of data-driven approaches that started already more than a decade ago, it is surprising at first that the pace of ML discoveries in medicine seems substantially slower than the ones found in consumer applications such as ChatGPT. The answer to that paradox is relatively simple: in healthcare, data is hard to…
-
Special Seminar: Anshul Kundaje, PhD – “Using deep learning models to debug regulatory genomics experiments and decode cis-regulatory syntax”
BWH Computational Pathology Special Seminar Title: Using deep learning models to debug regulatory genomics experiments and decode cis-regulatory syntax Speaker: Anshul Kundaje, PhD Affiliation: Stanford University Position: Associate Professor, Genetics […]
-
Nature Medicine Publications detail Mahmood Lab’s Design of AI Foundation Models to Advance Pathology
Two foundation models for pathology AI developed by the Mahmood Lab published in Nature Medicine: UNI and CONCH. Foundation models, advanced artificial intelligence systems trained on large-scale datasets, hold the […]